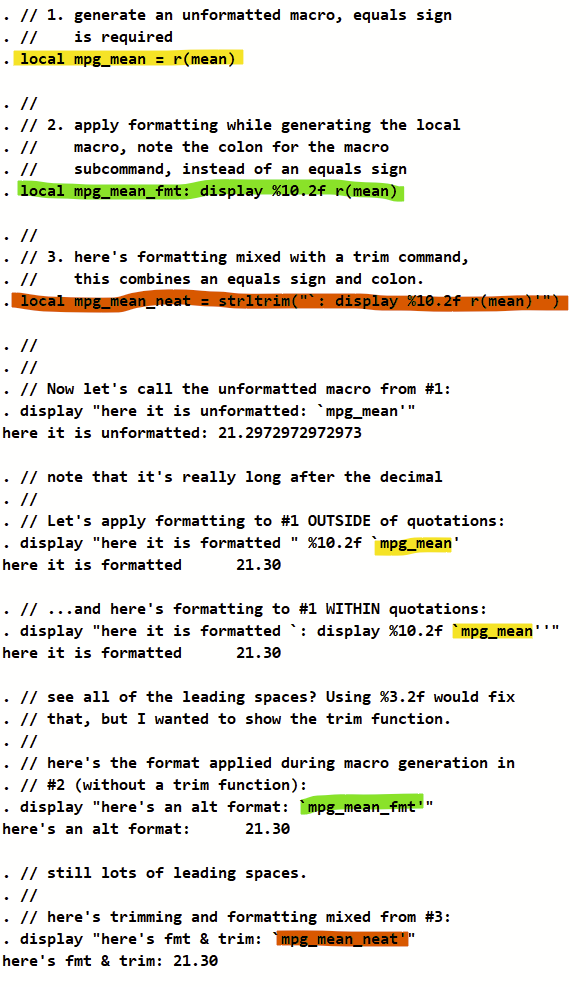
Mediation is a commonly-used tool in epidemiology. Inverse odds ratio-weighted (IORW) mediation was described in 2013 by Eric J. Tchetgen Tchetgen in this publication. It’s a robust mediation technique that can be used in many sorts of analyses, including logistic regression, modified Poisson regression, etc. It is also considered valid if there is an observed exposure*mediator interaction on the outcome.
There have been a handful of publications that describe the implementation of IORW (and its cousin inverse odds weighting, or IOW) in Stata, including this 2015 AJE paper by Quynh C Nguyen and this 2019 BMJ open paper by Muhammad Zakir Hossin (see the supplements of each for actual code). I recently had to implement this in a REGARDS project using a binary mediation variable and wanted to post my code here to simplify things. Check out the Nguyen paper above if you need to modify the following code to run IOW instead of IOWR, or if you are using a continuous mediation variable, rather than a binary one.
A huge thank you to Charlie Nicoli MD, Leann Long PhD, and Boyi Guo (almost) PhD who helped clarify implementation pieces. Also, Charlie wrote about 90% of this code so it’s really his work. I mostly cleaned it up, and clarified the approach as integrated in the examples from Drs. Nguyen and Hossin from the papers above.
IORW using pweight data (see below for unweighted version)
The particular analysis I was running uses pweighting. This code won’t work in data that doesn’t use weighting. This uses modified Poisson regression implemented as GLMs. These can be swapped out for other models as needed. You will have to modify this script if you are using 1. a continuous exposure, 2. more than 1 mediator, 3. a different weighting scheme, or 4. IOW instead of IORW. See the supplement in the above Nguyen paper on how to modify this code for those changes.
*************************************************
// this HEADER is all you should have to change
// to get this to run as weighted data with binary
// exposure using IORW. (Although you'll probably
// have to adjust the svyset commands in the
// program below to work in your dataset, in all
// actuality)
*************************************************
// BEGIN HEADER
//
// Directory of your dataset. You might need to
// include the entire file location (eg "c:/user/ ...")
// My file is located in my working directory so I just list
// a name here. Alternatively, can put URL for public datasets.
global file "myfile.dta"
//
// Pick a title for the table that will output at the end.
// This is just to help you stay organized if you are running
// a few of these in a row.
global title "my cool analysis, model 1"
//
// Components of the regression model. Outcome is binary,
// the exposure (sometimes called "dependent variable" or
// "treatment") is also binary. This code would need to be modified
// for a continuous exposure variable. See details in Step 2.
global outcome mi_incident
global exposure smoking
global covariates age sex
global mediator c.biomarker
global ifstatement if mi_prevalent==0 & biomarker<. & mi_incident <.
//
// Components of weighting to go into "svyset" command.
// You might have
global samplingweight mysamplingweightvar
global id id_num // ID for your participants
global strata mystratavar
//
// Now pick number of bootstraps. Aim for 1000 when you are actually
// running this, but when debugging, start with 50.
global bootcount 50
// and set a seed.
global seed 8675309
// END HEADER
*************************************************
//
//
//
// Load your dataset.
use "${file}", clear
//
// Drop then make a program.
capture program drop iorw_weighted
program iorw_weighted, rclass
// drop all variables that will be generated below.
capture drop predprob
capture drop inverseoddsratio
capture drop weight_iorw
//
*Step 1: svyset data since your dataset is weighted. If your dataset
// does NOT require weighting for its analysis, do not use this program.
svyset ${id}, strata(${strata}) weight(${samplingweight}) vce(linearized) singleunit(certainty)
//
*Step 2: Run the exposure model, which regresses the exposure on the
// mediator & covariates. In this example, the exposure is binary so we are
// using logistic regression (logit). If the exposure is a normally-distributed
// continuous variable, use linear regression instead.
svy linearized, subpop(${ifstatement}): logit ${exposure} ${mediator} ${covariates}
//
// Now grab the beta coefficient for mediator. We'll need that to generate
// the IORW weights.
scalar med_beta=e(b)[1,1]
//
*Step 3: Generate predicted probability and create inverse odds ratio and its
// weight.
predict predprob, p
gen inverseoddsratio = 1/(exp(med_beta*${mediator}))
//
// Calculate inverse odds ratio weights. Since our dataset uses sampling
// weights, we need to multiply the dataset's weights times the IORW for the
// exposure group. This step is fundamentally different for non-weighted
// datasets.
// Also note that this is for binary exposures, need to revise
// this code for continuous exposures.
gen weight_iorw = ${samplingweight} if ${exposure}==0
replace weight_iorw = inverseoddsratio*${samplingweight} if ${exposure}==1
//
*Step 4: TOTAL EFFECTS (ie no mediator) without IORW weighting yet.
// (same as direct effect, but without the IORW)
svyset ${id}, strata(${strata}) weight(${samplingweight}) vce(linearized) singleunit(certainty)
svy linearized, subpop(${ifstatement}): glm ${outcome} ${exposure} ${covariates}, family(poisson) link(log)
matrix bb_total= e(b)
scalar b_total=bb_total[1,1]
return scalar b_total=bb_total[1,1]
//
*Step 5: DIRECT EFFECTS; using IORW weights instead of the weighting that
// is used typically in your analysis.
svyset ${id}, strata(${strata}) weight(weight_iorw) vce(linearized) singleunit(certainty)
svy linearized, subpop(${ifstatement}): glm ${outcome} ${exposure} ${covariates}, family(poisson) link(log)
matrix bb_direct=e(b)
scalar b_direct=bb_direct[1,1]
return scalar b_direct=bb_direct[1,1]
//
*Step 6: INDIRECT EFFECTS
// indirect effect = total effect - direct effects
scalar b_indirect=b_total-b_direct
return scalar b_indirect=b_total-b_direct
//scalar expb_indirect=exp(scalar(b_indirect))
//return scalar expb_indirect=exp(scalar(b_indirect))
//
*Step 7: calculate % mediation
scalar define percmed = ((b_total-b_direct)/b_total)*100
// since indirect is total minus direct, above is the same as saying:
// scalar define percmed = (b_indirect)/(b_total)*100
return scalar percmed = ((b_total-b_direct)/b_total)*100
//
// now end the program.
end
//
*Step 8: Now run the above bootstraping program
bootstrap r(b_total) r(b_direct) r(b_indirect) r(percmed), seed(${seed}) reps(${bootcount}): iorw_weighted
matrix rtablebeta=r(table) // this is the beta coefficients
matrix rtableci=e(ci_percentile) // this is the 95% confidence intervals using the "percentiles" (ie 2.5th and 97.5th percentiles) rather than the default normal distribution method in stata. The rational for using percentiles rather than normal distribution is found in the 4th paragraph of the "analyses" section here (by refs 37 & 38): https://bmjopen.bmj.com/content/9/6/e026258.long
//
// Just in case you are curious, here are the the ranges of the 95% CI,
// realize that _bs_1 through _bs_3 need to be exponentiated:
estat bootstrap, all // percentiles is "(P)", normal is "(N)"
//
// Here's a script that will display your beta coefficients in
// a clean manner, exponentiating them when required.
quietly {
noisily di "${title} (bootstrap count=" e(N_reps) ")*"
noisily di _col(15) "Beta" _col(25) "95% low" _col(35) "95% high" _col(50) "Together"
local beta1 = exp(rtablebeta[1,1])
local low951 = exp(rtableci[1,1])
local high951 = exp(rtableci[2,1])
noisily di "Total" _col(15) %4.2f `beta1' _col(25) %4.2f `low951' _col(35) %4.2f `high951' _col(50) %4.2f `beta1' " (" %4.2f `low951' ", " %4.2f `high951' ")"
local beta2 = exp(rtablebeta[1,2])
local low952 = exp(rtableci[1,2])
local high952 = exp(rtableci[2,2])
noisily di "Direct" _col(15) %4.2f `beta2' _col(25) %4.2f `low952' _col(35) %4.2f `high952' _col(50) %4.2f `beta2' " (" %4.2f `low952' ", " %4.2f `high952' ")"
local beta3 = exp(rtablebeta[1,3])
local low953 = exp(rtableci[1,3])
local high953 = exp(rtableci[2,3])
noisily di "Indirect" _col(15) %4.2f `beta3' _col(25) %4.2f `low953' _col(35) %4.2f `high953' _col(50) %4.2f `beta3' " (" %4.2f `low953' ", " %4.2f `high953' ")"
local beta4 = (rtablebeta[1,4])
local low954 = (rtableci[1,4])
local high954 = (rtableci[2,4])
noisily di "% mediation" _col(15) %4.2f `beta4' "%" _col(25) %4.2f `low954' "%"_col(35) %4.2f `high954' "%" _col(50) %4.2f `beta4' "% (" %4.2f `low954' "%, " %4.2f `high954' "%)"
noisily di "*Confidence intervals use 2.5th and 97.5th percentiles"
noisily di " rather than default normal distribution in Stata."
noisily di " "
}
// the end.IORW for datasets that don’t use weighting (probably the one you are looking for)
Here is the code above, except without consideration of weighting in the overall dataset. (Obviously, IORW uses weighting itself.) This uses modified Poisson regression implemented as GLMs. These can be swapped out for other models as needed. You will have to modify this script if you are using 1. a continuous exposure, 2. more than 1 mediator, 3. a different weighting scheme, or 4. IOW instead of IORW. See the supplement in the above Nguyen paper on how to modify this code for those changes.
*************************************************
// this HEADER is all you should have to change
// to get this to run as weighted data with binary
// exposure using IORW.
*************************************************
// BEGIN HEADER
//
// Directory of your dataset. You might need to
// include the entire file location (eg "c:/user/ ...")
// My file is located in my working directory so I just list
// a name here. Alternatively, can put URL for public datasets.
global file "myfile.dta"
//
// Pick a title for the table that will output at the end.
// This is just to help you stay organized if you are running
// a few of these in a row.
global title "my cool analysis, model 1"
// Components of the regression model. Outcome is binary,
// the exposure (sometimes called "dependent variable" or
// "treatment") is also binary. This code would need to be modified
// for a continuous exposure variable. See details in Step 1.
global outcome mi_incident
global exposure smoking
global covariates age sex
global mediator c.biomarker
global ifstatement if mi_prevalent==0 & biomarker<. & mi_incident <.
//
// Now pick number of bootstraps. Aim for 1000 when you are actually
// running this, but when debugging, start with 50.
global bootcount 50
// and set a seed.
global seed 8675309
// END HEADER
*************************************************
//
//
//
// Load your dataset.
use "${file}", clear
//
// Drop then make a program.
capture program drop iorw
program iorw, rclass
// drop all variables that will be generated below.
capture drop predprob
capture drop inverseoddsratio
capture drop weight_iorw
//
//
*Step 1: Run the exposure model, which regresses the exposure on the
// mediator & covariates. In this example, the exposure is binary so we are
// using logistic regression (logit). If the exposure is a normally-distributed
// continuous variable, use linear regression instead.
logit ${exposure} ${mediator} ${covariates} ${ifstatement}
//
// Now grab the beta coefficient for mediator. We'll need that to generate
// the IORW weights.
scalar med_beta=e(b)[1,1]
//
*Step 2: Generate predicted probability and create inverse odds ratio and its
// weight.
predict predprob, p
gen inverseoddsratio = 1/(exp(med_beta*${mediator}))
//
// Calculate inverse odds ratio weights.
// Also note that this is for binary exposures, need to revise
// this code for continuous exposures.
gen weight_iorw = 1 if ${exposure}==0
replace weight_iorw = inverseoddsratio if ${exposure}==1
//
*Step 3: TOTAL EFFECTS (ie no mediator) without IORW weighting yet. (same as direct effect, but without the IORW)
glm ${outcome} ${exposure} ${covariates} ${ifstatement}, family(poisson) link(log) vce(robust)
matrix bb_total= e(b)
scalar b_total=bb_total[1,1]
return scalar b_total=bb_total[1,1]
//
*Step 4: DIRECT EFFECTS; using IORW weights
glm ${outcome} ${exposure} ${covariates} ${ifstatement} [pweight=weight_iorw], family(poisson) link(log) vce(robust)
matrix bb_direct=e(b)
scalar b_direct=bb_direct[1,1]
return scalar b_direct=bb_direct[1,1]
//
*Step 5: INDIRECT EFFECTS
// indirect effect = total effect - direct effects
scalar b_indirect=b_total-b_direct
return scalar b_indirect=b_total-b_direct
//scalar expb_indirect=exp(scalar(b_indirect))
//return scalar expb_indirect=exp(scalar(b_indirect))
//
*Step 6: calculate % mediation
scalar define percmed = ((b_total-b_direct)/b_total)*100
// since indirect is total minus direct, above is the same as saying:
// scalar define percmed = (b_indirect)/(b_total)*100
return scalar percmed = ((b_total-b_direct)/b_total)*100
//
// now end the program.
end
//
*Step 7: Now run the above bootstraping program
bootstrap r(b_total) r(b_direct) r(b_indirect) r(percmed), seed(${seed}) reps(${bootcount}): iorw
matrix rtablebeta=r(table) // this is the beta coefficients
matrix rtableci=e(ci_percentile) // this is the 95% confidence intervals using the "percentiles" (ie 2.5th and 97.5th percentiles) rather than the default normal distribution method in stata. The rational for using percentiles rather than normal distribution is found in the 4th paragraph of the "analyses" section here (by refs 37 & 38): https://bmjopen.bmj.com/content/9/6/e026258.long
//
// Just in case you are curious, here are the the ranges of the 95% CI,
// realize that _bs_1 through _bs_3 need to be exponentiated:
estat bootstrap, all // percentiles is "(P)", normal is "(N)"
//
// Here's a script that will display your beta coefficients in
// a clean manner, exponentiating them when required.
quietly {
noisily di "${title} (bootstrap count=" e(N_reps) ")*"
noisily di _col(15) "Beta" _col(25) "95% low" _col(35) "95% high" _col(50) "Together"
local beta1 = exp(rtablebeta[1,1])
local low951 = exp(rtableci[1,1])
local high951 = exp(rtableci[2,1])
noisily di "Total" _col(15) %4.2f `beta1' _col(25) %4.2f `low951' _col(35) %4.2f `high951' _col(50) %4.2f `beta1' " (" %4.2f `low951' ", " %4.2f `high951' ")"
local beta2 = exp(rtablebeta[1,2])
local low952 = exp(rtableci[1,2])
local high952 = exp(rtableci[2,2])
noisily di "Direct" _col(15) %4.2f `beta2' _col(25) %4.2f `low952' _col(35) %4.2f `high952' _col(50) %4.2f `beta2' " (" %4.2f `low952' ", " %4.2f `high952' ")"
local beta3 = exp(rtablebeta[1,3])
local low953 = exp(rtableci[1,3])
local high953 = exp(rtableci[2,3])
noisily di "Indirect" _col(15) %4.2f `beta3' _col(25) %4.2f `low953' _col(35) %4.2f `high953' _col(50) %4.2f `beta3' " (" %4.2f `low953' ", " %4.2f `high953' ")"
local beta4 = (rtablebeta[1,4])
local low954 = (rtableci[1,4])
local high954 = (rtableci[2,4])
noisily di "% mediation" _col(15) %4.2f `beta4' "%" _col(25) %4.2f `low954' "%"_col(35) %4.2f `high954' "%" _col(50) %4.2f `beta4' " (" %4.2f `low954' ", " %4.2f `high954' ")"
noisily di "*Confidence intervals use 2.5th and 97.5th percentiles"
noisily di " rather than default normal distribution in Stata."
noisily di " "
}
// the end.