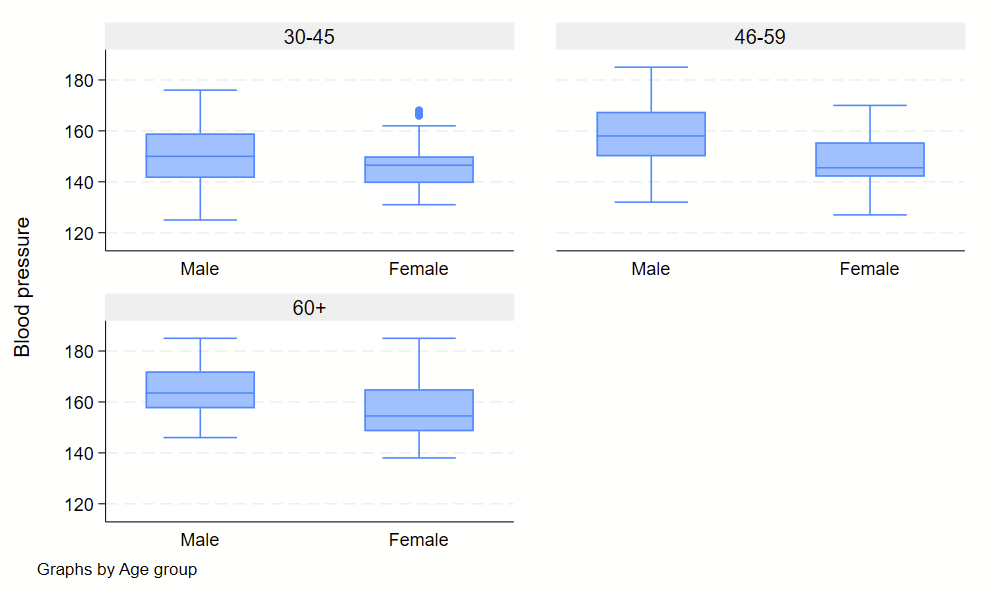
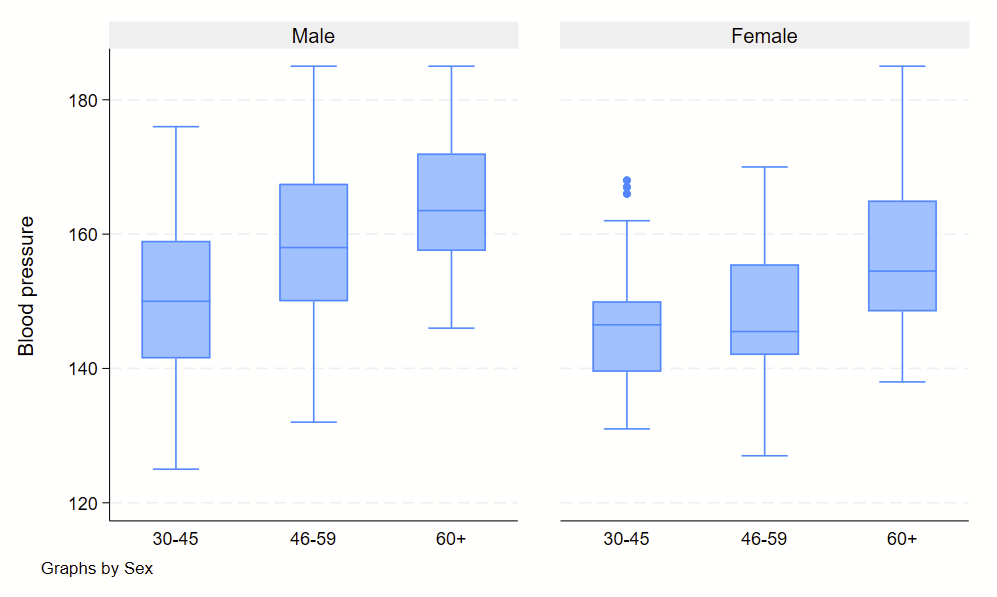
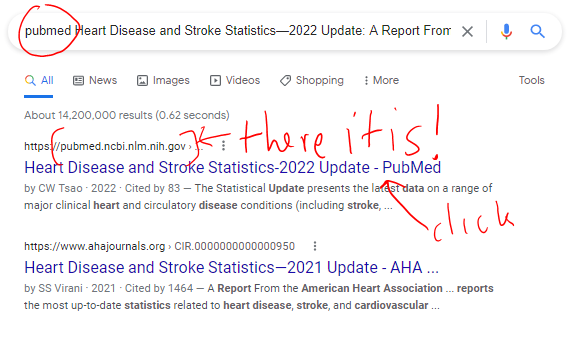
There are all sorts of models out there for performing regressions. This page focuses on 3 that are used for binary outcomes:
- Logistic regression
- Modified Poisson regression
- Cox proportional hazards models.
Getting set up
Before you get going, you want to explicitly define your outcome of interest (aka dependent variable), primary exposure (aka independent variable) and covariates that you are adjusting for in your model (aka other independent variables). You’ll also want to know your study design (eg case-control? cross-sectional? prospective? time to event?).
Are your independent variables continuous or factor/categorical?
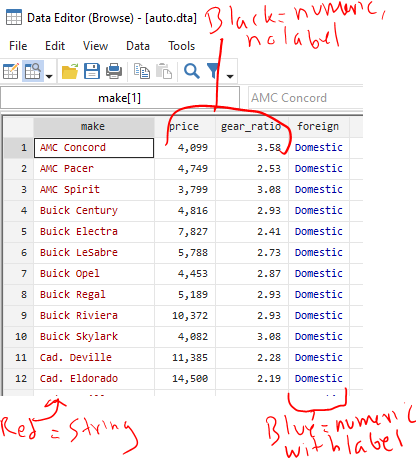
In these regression models, you can specify that independent variables (primary exposure and covariates that are in your model) are continuous or factor/categorical. Continuous variables can be specified with a leading “c.”, and examples might include age, height, or length. (“c.age”.) In contrast, factor/categorical variables might be New England states (e.g., 1=Connecticut, 2=Maine, 3=Mass, 4=New Hampshire, 5=RI, and 6=Vermont). So, you’d want to specifiy a variable for “newengland” as i.newengland in your regression. Stata defaults to treating the smallest number (here, 1=Connecticut) as the reference group. You can change that by using the “ib#.” prefix instead, where the # is the reference group, or here ib2.newengland to have Maine as the reference group.
Read about factor variables in –help fvvarlist–.
What about binary/dichotomous variables (things that are ==0 or 1)? Well, it doesn’t change your analysis if you specify a binary/dichotomous as either continuous (“c.”) or factor/categorical (“i.”). The math is the same on the back end.
Checking for interactions
In general, when checking for an interaction, you will need to specify if the two variables of interest are categorical or factor/categorical and drop two pound signs in between the two. See details on –help fvvarlist–. Here’s an example of how this would look, looking for an interaction between sex on age groups.
regress bp i.sex##c.agegrp
Source | SS df MS Number of obs = 240
-------------+---------------------------------- F(3, 236) = 23.86
Model | 9519.9625 3 3173.32083 Prob > F = 0.0000
Residual | 31392.8333 236 133.02048 R-squared = 0.2327
-------------+---------------------------------- Adj R-squared = 0.2229
Total | 40912.7958 239 171.183246 Root MSE = 11.533
------------------------------------------------------------------------------
bp | Coefficient Std. err. t P>|t| [95% conf. interval]
-------------+----------------------------------------------------------------
sex |
Female | -4.275 3.939423 -1.09 0.279 -12.03593 3.485927
agegrp | 7.0625 1.289479 5.48 0.000 4.52214 9.60286
|
sex#c.agegrp |
Female | -1.35 1.823599 -0.74 0.460 -4.942611 2.242611
|
_cons | 143.2667 2.785593 51.43 0.000 137.7789 148.7545
------------------------------------------------------------------------------
You’ll see the sex#c.agegrp P-value is 0.460, so that wouldn’t qualify as a statistically significant interaction.
Regressions using survey data?
If you are using survey analyses (eg need to account for pweighting), generally you have to use the svy: prefixes for your analyses. This includes svy: logistic, poisson, etc. Type –help svy_estimation– to see what the options are.
Logistic regression
Logistic regressions provide odds ratios of binary outcomes. Odds ratios don’t approximate the risk ratio if the outcome is common (eg >10% occurrence) so I tend to avoid them as I study hypertension, which occurs commonly in a population.
There are oodles of details on logistic regression in the excellent UCLA website. In brief, you want want to use a regression command, you can use “logit” and get the raw betas or “logistic” and get the odds ratios. Most people will want to use “logistic” to get the odds ratios.
Here’s an example of logistic regression in Stata. In this dataset, “race” is Black/White/other, so you’ll need to specify this as a factor/categorical variable with the “i.” prefix, however “smoke” is binary so you can either specify it as a continuous or as a factor/categorical variable. If you don’t specify anything, then it is treated as continuous, which is fine.
webuse lbw, clear
codebook race // see race is 1=White, 2=Black, 3=other
codebook smoke // smoke is 0/1
logistic low i.race smoke
// output:
Logistic regression Number of obs = 189
LR chi2(3) = 14.70
Prob > chi2 = 0.0021
Log likelihood = -109.98736 Pseudo R2 = 0.0626
------------------------------------------------------------------------------
low | Odds ratio Std. err. z P>|z| [95% conf. interval]
-------------+----------------------------------------------------------------
race |
Black | 2.956742 1.448759 2.21 0.027 1.131716 7.724838
Other | 3.030001 1.212927 2.77 0.006 1.382616 6.64024
|
smoke | 3.052631 1.127112 3.02 0.003 1.480432 6.294487
_cons | .1587319 .0560108 -5.22 0.000 .0794888 .3169732
------------------------------------------------------------------------------
Note: _cons estimates baseline odds.
So, here you see that the odds ratio (OR) for the outcome of “low” is 2.96 (95% CI 1.13, 7.72) for Black relative to White participants and the OR for the outcome of “low” is 3.03 (95% CI 1.38, 6.64) for Other race relative to White participants. Since it’s the reference group, you’ll notice that White race isn’t shown. But what if we want Black race as the reference? You’d use “ib2.race” instead of “i.race”. Example:
logistic low ib2.race smoke
// output:
Logistic regression Number of obs = 189
LR chi2(3) = 14.70
Prob > chi2 = 0.0021
Log likelihood = -109.98736 Pseudo R2 = 0.0626
------------------------------------------------------------------------------
low | Odds ratio Std. err. z P>|z| [95% conf. interval]
-------------+----------------------------------------------------------------
race |
White | .3382101 .1657178 -2.21 0.027 .1294525 .8836136
Other | 1.024777 .5049663 0.05 0.960 .3901157 2.691938
|
smoke | 3.052631 1.127112 3.02 0.003 1.480432 6.294487
_cons | .4693292 .2059043 -1.72 0.085 .1986269 1.108963
------------------------------------------------------------------------------
Note: _cons estimates baseline odds.
Modified Poisson regression
Modified Poisson regression (sometimes called Poisson Regression with Robust Variance Estimation or Poisson Regression with Sandwich Variance Estimation) is pretty straightforward. Note: this is different from ‘conventional’ Poisson regression, which is used for counts and not dichotomous outcomes. You use the “poisson” command with subcommands “, vce(robust) irr” to use the modified Poisson regression type. Note: with svy data, robust VCE is the default so you just need to use the subcommand “, irr”.
As with the logistic regression section above, race is a 3-level nominal variable so we’ll use the “i.” or “ib#.” prefix to specify that it’s not to be treated as a continuous variable.
webuse lbw, clear
codebook race // see race is 1=White, 2=Black, 3=other
codebook smoke // smoke is 0/1
// set Black race as the reference group
poisson low ib2.race smoke, vce(robust) irr
// output:
Iteration 0: Log pseudolikelihood = -122.83059
Iteration 1: Log pseudolikelihood = -122.83058
Poisson regression Number of obs = 189
Wald chi2(3) = 19.47
Prob > chi2 = 0.0002
Log pseudolikelihood = -122.83058 Pseudo R2 = 0.0380
------------------------------------------------------------------------------
| Robust
low | IRR std. err. z P>|z| [95% conf. interval]
-------------+----------------------------------------------------------------
race |
White | .507831 .141131 -2.44 0.015 .2945523 .8755401
Other | 1.038362 .2907804 0.13 0.893 .5997635 1.7977
|
smoke | 2.020686 .4300124 3.31 0.001 1.331554 3.066471
_cons | .3038099 .0800721 -4.52 0.000 .1812422 .5092656
------------------------------------------------------------------------------
Note: _cons estimates baseline incidence rate.
Here you see that the relative risk (RR) of “low” for White relative to Black participants is 0.51 (95% CI 0.29, 0.88), and the RR for other race relative to Black participants is 1.04 (95% CI 0.60, 1.80). As above, the “Black” group isn’t shown since it’s the reference group.
Cox proportional hazards model
Cox PH models are used for time to event data. This is a 2-part. First is to –stset– the data, thereby telling Stata that it’s time to event data. In the –stset– command, you specify the outcome/failure variable. Second is to use the –stcox– command to specify the primary exposure/covariates of interest (aka independent variables). There are lots of different steps to setting up the stset command, so make sure to check the –help stset– page. Ditto –help stcox–.
Here, you specify the days of follow-up as “days_hfhosp” and failure as “hfhosp”. Since follow-up is in days, you set the scale to 365.25 to have it instead considered as years.
stset days_hfhosp, f(hfhosp) scale(365.25)Then you’ll want to use the stcox command to estimate the risk of hospitalization by beta blocker use by age, sex, and race with race relative to the group that is equal to 1.
stcox betablocker age sex ib1.raceYou’ll want to make a Kaplan Meier figure at the same time, read about how to do that on this other post.