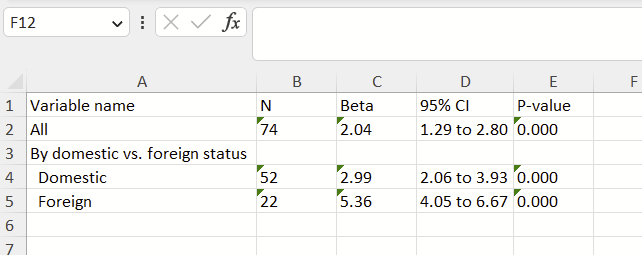
Everyone knows their (5-digit) ZIP and it can be linked to population-level data. ZIP Codes have limitations since they were designed for mail delivery and not for population details. You can easily get county data from these data as well.
In epidemiological studies (especially EMR and survey data), you’ll almost certainly have a ZIP code or county, and almost never a census tract. It’s easy to find census data sets, but finding the analogous ZIP code dataset is a bit tricker. Every time I try to do a project with ZIP codes, I kick myself for not keeping a list of ZIP code data sets. So, this page will keep a running list of ZIP code-linked datasets. It’ll be updated periodically. If there’s a useful resource that I have missed, please email me at timothy.plante@uvm.edu and I’ll add it.
A few technical notes:
- US Postal Service (USPS) ZIP codes – It seems that some datasets use a variation of USPS’s active ZIP codes. These are constantly being updated by the US Postal Service. ZIP codes are either the ‘standard’ 5-digit ZIP code or ZIP+4 (e.g., 9 digit). You can narrow down a lot further with the ZIP+4 version, but often times you only have the 5-digit ZIP.
- ZCTA stands for ZIP Code Tabulation Area and is the US Census’s take on representing the topology of ZIP codes. These are produced for the q10y census. There are different ZCTAs for the 2000 Census and 2010 Census (as of 12/2020). Details about the US Census’s approach to developing ZCTAs can be found here.
- You can read about the differences between ZCTA and USPS ZIP codes here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1762013/
- The 2010 ZCTA to county, subdivision, tract, congressional district, metropolitan and micropolitan statistical area, and New England City and Town Area can be found here.
- Note, there is also a non-ZCTA 5-digit ZIP code standard used by the US Census are specific to the ZIP Code Business Patterns Survey. So, these relate to businesses, not people. Details are here.
- USPS ZIP code to ZCTA crosswalk – this is provided by UDS at this website: https://udsmapper.org/zip-code-to-zcta-crosswalk/
- The US Housing and Urban Development (HUD) also has its own ZIP linkage, which can be found here. You can read about the details of the HUD ZIP crosswalk here. This is not the same as ZCTA. The nice thing about the HUD ZIP crosswalk is that it’s updated quarterly, it links to 2000 or 2010 US Census county or tract GEOID via FIPS code, and the OMB’s core-based statistical area (CBSA; basically definitions of urban groups), and congressional districts. It also provides some details about residential vs. business vs. other addresses in that zip code.
Linking ZIP to county and FIPS
There’s a dataset on the HUD website here, on the “select crosswalk type” dropdown, select ZIP-County. From my read, this is ZCTA ZIP code for more recent datasets, but that isn’t explicitly stated. There’s also this resource from Dan Ofer that I haven’t had the chance to use but looks promising.
Cartography
Here are some resources if you want to make a map.
US Census (ZCTA)
Here’s some mapping files provided by the US Census.
Here’s a great Stata-specific page with both the ZCTA and US Postal Service ZIP files. I recommend the ZCTA if you will be using US census data.
HUD-ZIP linkage
Details are here.
USPS ZIP code
Here’s a commonly-used dataset from Esri’s ArcGIS.
Here’s the USPS ZIP code for Stata.
Distance between zip codes
Can be found here from the NBER here.
Health outcomes, prevention, health risk behaviors, disability, health status, and SDOH at CDC Places/500 Cities (ZCTA)
The RWJF/CDC 500 cities and Places database provides a huge collection of data linked to ZIP, with data originating from BRFSS and ACS/Census. Data can be downloaded at CDC places (ZCTA).
Commercial datasets
You might want to take a look at datasets available, some for free, some for a fee, on the AWS marketplace here (this is a link to the search term “Zip”). There are at least a few companies that sell datasets with things like demographics for Zip+4 (eg V12 on AWS), which you might be interested in. While you’re at it, you might also opt to look at Census files on this marketplace here.
Demographics
US Census (ZCTA)
The US Census used to distribute their summary files via FTP for their 2000 census and 2010 census. [Note: those are links to the Summary File 1, which doesn’t include rurality. Those are in Summary File 2.] These 39 and 47 files that must be merged by some convoluted process that I’m not going to try to figure out. Fortunately, the National Bureau of Economic Research (NBER) generated Summary File 1-ZCTA linked files for Stata, SAS, and CSV files that can be downloaded here:
As an example of how to use the NBER files, let’s look at the 2010 files. Files are indexed in this Census Summary File 1 (SF1) document. Search for “File 03” in that PDF to find the details for File 03 on page 184. Note that “P0030002” through “P0030008” are variables for race in the entire population. File 04 then has race and ethnicity among adults (male sex is “P0120002”, female sex is “P0120026”). File 07 has sex by race/ethnicity and age, and so on. You’ll want to save the specific variables from each of these files and generate your own dataset, depending on what you are attempting to do.
But what about rurality? That’s in the Summary File 2 (SF2) document. The US Census data used to be on a website called American Fact Finder, which was simple to use and wasn’t annoying. More recently it was moved to data.census.gov, which is a spiffy looking website that is in all actuality, quite terrible and I want it to go away. I can’t figure out how to download what I want. I tried to make a walkthrough of how to download urban/rurality by ZCTA but it gave me a blank table. Fortunately, I had downloaded it from American Fact Finder before it went offline. You can download the version that I saved here.
An alternative to data.census.gov in the wake of the loss of American Fact Finder is the NHGIS website. You can also try IPUMS, which also has data related to global health, GIS, time use, health surveys (NHIS and MEPS), and higher education information. Finally, there are a whole host of free Census files on AWS, I haven’t tried these but you might want to take a look here.
Social Determinants of Health
Take a look at this handy review of SDoH definitions by Lori Dean and Roland Thorpe at JHU. Before I go any further, I recommend looking at the PhenX toolkit for their resources on SDoH. There are a few conceptual frameworks for SDOH. Here’s Healthy People 2030. Here’s the WHO one. I like the KFF’s Figure 1 here, which defines the following factors (note there’s plenty of overlap between Healthy People 2030 and KFF). Here’s a working list of resources that I’ll keep adding to, organized by KFF’s figure 1 overview:
- Economic stability – Employment, income, expenses, debt, medical bills, support.
- Neighborhood and physical environment – Housing, transportation, safety, parks, playgrounds, walkability, ZIP code/geography.
- Walk Score, a commercial (not free) database, can be linked to ZIP code it seems: https://www.walkscore.com/professional/research.php
- Can apply RUCA codes or rural scores for counties. See “Rurality” section below.
- US Forestry Tree Canopy (I haven’t downloaded to confirm it uses ZIP, but it appears to have GIS linkage at the least) https://data.fs.usda.gov/geodata/rastergateway/treecanopycover/index.php
- Alternatively, the MODIS Vegetation Index’s normalized difference vegetation index (NDVI) and enhanced vegetation index (EVI) are available from NASA. Not by Zip but by 1-km overview of the US it seems. https://modis.gsfc.nasa.gov/data/dataprod/mod13.php
- EPA ecoregions, with GIS (not ZIP) data https://www.epa.gov/eco-research/ecoregions-north-america
- Pollution metrics for census tracts, block groups, county, state, and national. Not zip but this is awesome. Ozone (O3), CO, SO2, NO2, PM10, and PM2.5 available here: https://www.caces.us/
- EPA Toxics Release inventory: https://www.epa.gov/toxics-release-inventory-tri-program
- HUD’s fair market rent cost, by ZIP: https://www.huduser.gov/portal/datasets/fmr/smallarea/index.html (H/t to the Data Is Plural email.)
- USPS’s change of address by ZIP, under “Frequently requested records”): https://about.usps.com/who/legal/foia/library.htm (H/t to the Data Is Plural email.)
- Crowding, housing cost burden, broadband use at CDC places (ZCTA)
- Education – Literacy, language, early childhood education, vocational training, higher education.
- Food – Hunger, access to healthy options.
- Community and social context – Social integration, support systems, community engagement, discrimination, stress.
- Health care system – Health coverage, provider availability, provider linguistic and cultural competency, quality of care.
- The National Health Care Survey (NCHS) from the CDC details a huge amount about the American healthcare system. it has ZIP-code level information in their restricted use dataset. A list of items is available here: https://www.cdc.gov/rdc/geocodes/geowt_nhcs.htm …and details about the NCHS are here: https://www.cdc.gov/nchs/dhcs/index.htm
- Can define with via the Health Professional Shortage Area (HPSA) metric from HRSA, I’ll come back and add details later.
- Can also apply metrics for public health using the County Health Rankings (see below).
- Other
- Social capital atlas – with economic connectedness (how much low- and high-income people are friends), cohesiveness (degree to which social networks are fragmented into cliques), and civic engagement (rates of volunteering and participating in community organization, with data by zip, county, colleges, and high schools: https://socialcapital.org/
As I expand these, I will do my best to cover as many of these as possible, as how they apply to ZIP code and county.
Social Deprivation Index or SDI (ZCTA)
Derived from the American Community Survey 5-year estimates. Details include overall SDI score, income, education, employment, housing (% living in crowded rentals), household characteristics (% of single parent households with dependents who are minors), transportation (% car non-ownership), demographics (% black population, % high needs population). Details and download files can be found here.
Here’s the original description, prior to the use of ZCTA. This manuscript only discusses the Primary Care Service Areas (PCSAs), from the Dartmouth Atlas: https://pubmed.ncbi.nlm.nih.gov/22816561/
Area Deprivation Index or ADI (ZIP+4)
More to come. Download site is here. You need to make a free account to access the data. You have to download each state individually, as an FYI.
HUD datasets on housing, income, etc. (can use the HUD-ZIP crosswalk)
Here is the website: https://hudgis-hud.opendata.arcgis.com/
I haven’t explored these data files much, but some details are below. The only file that natively includes the ZCTA is the Difficult Development Areas, under Community Development below.
- Agency administration – How the HUD is divided. Yawn.
- Community development – Community development block grant, LIHTC Qualified Census Tracts (aka low income), Difficulty Development Areas for Low Income Housing Tax Credit (LIHTC; high cost of living relative to Area Median Gross Income; interestingly using the ZCTA for metropolitan areas), Neighborhood Stabilization Program (purchase of abandoned buildings), Empowerment Zone/Enterprise Community/Renewal Community (economic growth tax incentives), Revitalization Areas.
- Community indicators – Details by American Community Survey, self-reported perceived rural/urban status (see Rurality section below), low-to-moderate income population by tract from the American Community Survey, Location Affordability Index from the American Community Survey, extreme temperatures by 1 degree latitude and longitude.
- Fair housing – More to come.
- Housing counseling – More to come.
- Initiatives and demonstrations – More to come.
- Location affordability – More to come.
- Mortgage insurance programs – More to come.
- Rental assistance programs – More to come.
- Disaster recovery – More to come.
Rurality
RUCA codes (Unclear ZIP type)
There was a bug in the 2010 US Census-derived RUCA-ZIP and the linkage was updated in 2020, and can be found here. I’m trying to figure out whether RUCA is most similar to ZCTA or USPS ZIP Codes. I’ll come back and update what I find out. Update: I didn’t get a response to my inquiry. Since this is linked to the Census data, it’s possibly ZCTA.
American Housing Survey (AHS) from HUD
Urbanization Perceptions Small Area Index. This was self-reported neighborhood as urban, suburban, or rural. Link is here.
US Census (ZCTA)
The US Census details their take on rurality here. The actual rurality details for the 2010 census are in “Summary File 2”, details of which can be found here. As documented above, data.census.gov is a barrier to downloading census data. Fortunately, I grabbed rurality by ZCTA from American Fact Finder before it was shut down. You can grab my file here.
NCHS Urban-Rural Classification (Counties)
This is a very popular classification methodology people frequently use this scheme so I’m including it here. Details are here.
Health data
County health rankings (county)
Much county-level data can be obtained from the excellent County Health Rankings website from UWI, sponsored by RWJF. These include “ranked” and “unranked” data, the sources of these datapoints are listed in the Excel files that you can download on the website (eg, Vermont’s is here). Ranked includes premature death (deaths <75y), poor fair health, poor physical health, poor mental health, low birthweight, adult smoking adult obesity, food environment index, physical inactivity, access to exercise opportunities, excessive drinking alcohol-impaired driving deaths, STIs, teen births, % uninsured <65, ratio of population to PCPs, ratio of population to dentists, preventable health stays, mammography screening, flu vaccinations, level of education, unemployment, % children in poverty, income inequality, children in single-parent households, social associations, violent crime, injury deaths, air pollution by particulate matter, drinking water violations, households with overcrowding/high housing costs/lack of kitchen facilities/lack of plumbing facilities, % that drive to work alone, long commutes. Unranked includes life expectancy, premature age-adjusted mortality, child mortality, infant mortality, quality of life metrics (frequent physical distress, frequent mental distress, diabetes and HIV prevalence), food insecurity, limited access to healthy foods, drug overdose deaths, motor vehicle crash deaths, insufficient sleep, uninsured adults, uninsured children, ratio of population to primary care providers, disconnected youth (% of 16-19 yo not in school or working), reading scores, math scores, median income, % children eligible for free or reduced price lunch, residential segregation, homicides, suicidies, ,firearm fatalities, juvenile arrests, traffic volume, home ownership, severe housing cost burden, and specific census details.
I can’t find a “download all” option, but the datasets use a preserved naming structure in the download directory, so if you copy the link for one state, you can replace it with the name for another state (replacing spaces with percent sign 20 if spaces if needed) to get that download. It’d be easy to build a loop in Stata to automate the download for all of these datasets.
Ecology
The Ecolo-Zip project includes global postal codes and US ZIP codes to provide “Combining two large-scale satellite image resources (ASTER; SRTM) and a customised customized geospatial sampling model, we provide high-resolution indicators of physical topography (elevation, mountainousness, distance to sea), vegetation (normalized difference vegetation index) and climate (surface temperature). A cross-validation between ASTER and SRTM elevation data demonstrated high concordance (ICC = 0.999).” H/t to the Data Is Plural email for this head’s up.
Other
CBSA – Core-based statistical area
The White House’s OMB defines the CBSA, which is broadly metropolitan areas. So NYC has NYC itself as well as the suburban areas of NYC (NJ, Westchester, etc.) HUD provides USPS ZIP crosswalk here.